Advanced data manipulation with tidyr and its allies
Jim Félix-Faure
October 2020
- Change table format
- Split two values from one cell
- Data set connection
- Manipulation of NA
- Practice
- Useful links
Note : To follow this workshop it is recommended to have already completed the following courses Fast data manipulation with dplyr and its allies and Rapid data visualization with ggplot2 (available on the Numerilab website - see link at the bottom of this document) or to have some basics on the use of the pipe operator, ggplot2 and the basic functions of the tidyverse.
Change table format
To optimize processing and/or visualization, it is recommended that the data tables under R be organized as follows :
- Each column is a variable..
- Each row is an observation.
- Each cell is composed of one value.
Let’s see some examples where this is not the case, and the solutions to apply using the tidyr package, included with the tidyverse.
From wide format to longer format
Here is an example where values are hidden in column names.
The weight of observations A and B is reported according to the year.
library(tidyverse)
Tbl_Weight <- tibble(
Observation = c("A", "B"),
"2010" = c(3, 3),
"2011" = c(5, 2),
"2012" = c(4, 3),
"2013" = c(5, 3)
)
Tbl_Weight
# A tibble: 2 x 5
Observation `2010` `2011` `2012` `2013`
<chr> <dbl> <dbl> <dbl> <dbl>
1 A 3 5 4 5
2 B 3 2 3 3
How to compute the average weight of each observation? How to draw a graph representing the weight of each observation over time?
Not so easy, unless you reorganize the data. Here the objective is twofold:
- create a variable
Yeargrouping the years present in the name of the columns. - create a variable
Weightgrouping the weight values keeping the correspondence with the years.
Tbl_WeightTidy <- Tbl_Weight %>%
pivot_longer(
cols = !Observation, # Columns to be used
names_to = "Year", # Variable where the column names will be stored
names_transform = list(Year = as.integer), # Choice of variable type for names_to
values_to = "Weight" # Variable where the values will be stored
)
Tbl_WeightTidy
# A tibble: 8 x 3
Observation Year Weight
<chr> <int> <dbl>
1 A 2010 3
2 A 2011 5
3 A 2012 4
4 A 2013 5
5 B 2010 3
6 B 2011 2
7 B 2012 3
8 B 2013 3
The average weight of observations :
Tbl_WeightTidy %>%
group_by(Observation) %>%
summarise(Mean = mean(Weight),
Sd = sd(Weight),
n = n())
`summarise()` ungrouping output (override with `.groups` argument)
# A tibble: 2 x 4
Observation Mean Sd n
<chr> <dbl> <dbl> <int>
1 A 4.25 0.957 4
2 B 2.75 0.5 4
Graph of weights as a function of years :
Tbl_WeightTidy %>%
ggplot() +
geom_line(aes(x = Year, y = Weight, color = Observation))
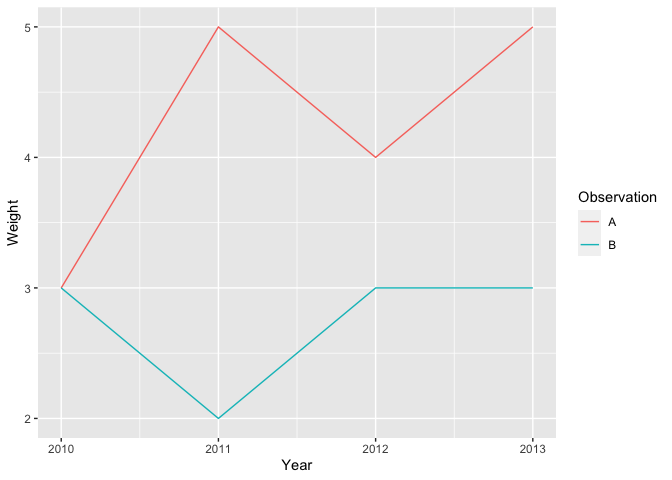
From large format to wider format
Sometimes the problem is reversed. One observation is present in several rows and several variables are in the same column.
Tbl_Water <- tibble(
Observation = rep(c("A", "B", "C", "D"), each = 2),
Measure = rep(c("pH", "O2"), 4),
Value = c(6, 99, 7, 90, 7.2, 85, 6.5, 96)
)
Tbl_Water
# A tibble: 8 x 3
Observation Measure Value
<chr> <chr> <dbl>
1 A pH 6
2 A O2 99
3 B pH 7
4 B O2 90
5 C pH 7.2
6 C O2 85
7 D pH 6.5
8 D O2 96
Again, it is not easy to graph the relationship between pH and O2 and the units in the Value column are not clear.
Tbl_WaterTidy <- Tbl_Water %>%
pivot_wider(names_from = Measure, # column that contains the variable names
values_from = Value # column that contains the values
)
Tbl_WaterTidy
# A tibble: 4 x 3
Observation pH O2
<chr> <dbl> <dbl>
1 A 6 99
2 B 7 90
3 C 7.2 85
4 D 6.5 96
Tbl_WaterTidy %>%
ggplot(aes(x = pH, y = O2)) +
geom_point()
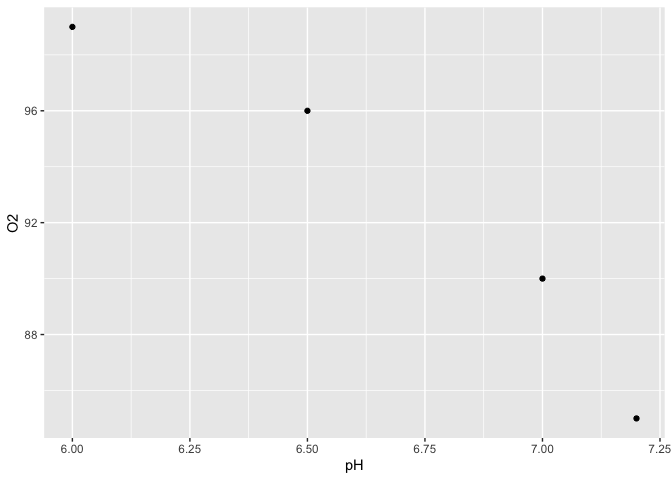
Split two values from one cell
When entering data, it can happen that two values are put together in the same cell.
For example, here are the results of the number of animals positive to a molecule on the number of animals tested.
Tbl_Effect <- tibble(
Molecule = c("A","B","C"),
Result = c("87/112","23/48","34/89")
)
Tbl_Effect
# A tibble: 3 x 2
Molecule Result
<chr> <chr>
1 A 87/112
2 B 23/48
3 C 34/89
We want to calculate the ratio between positive and tested animals.
The separate() function allows you to split the contents of one character column into several other character or numeric columns. It cuts out every character that is neither a number nor a letter. If necessary, the separator can be specified manually with the argument sep="".
Tbl_Effect %>%
separate(
col = Result, # which column to separate
into = c("Positive_animals", "Tested_animals"), # how to call the new columns
convert = TRUE, # convert the new columns to the appropriate type
remove = TRUE # remove the former column
) %>%
mutate(Ratio = Positive_animals/Tested_animals)
# A tibble: 3 x 4
Molecule Positive_animals Tested_animals Ratio
<chr> <int> <int> <dbl>
1 A 87 112 0.777
2 B 23 48 0.479
3 C 34 89 0.382
Data set connection
The joins
As the number, source, and diversity of the data increase, it is common to have multiple data tables. In this case, it is possible to connect them using a key. Its nature can vary (character, factor, numeric…) but it is essential that this key be a unique value for each observation.
Let’s take a look at an example:
Tbl_Site <- tibble(
Site = c("356a", "da4b", "77de", "1b64"),
Landuse = c("Forest", "Forest", "Pasture", "Wetland"),
Area = c(12, 30, 8, 17)
)
Tbl_Site
# A tibble: 4 x 3
Site Landuse Area
<chr> <chr> <dbl>
1 356a Forest 12
2 da4b Forest 30
3 77de Pasture 8
4 1b64 Wetland 17
Tbl_Result <- tibble(
Site = c("1b64", "1b64", "356a", "da4b", "da4b", "abcd"),
Measure = c("pH", "pH", "Corg", "pH", "Corg", "Corg"),
Value = c(7, 7.2, 3.5, 5.8, 2.7, 5.8)
)
Tbl_Result
# A tibble: 6 x 3
Site Measure Value
<chr> <chr> <dbl>
1 1b64 pH 7
2 1b64 pH 7.2
3 356a Corg 3.5
4 da4b pH 5.8
5 da4b Corg 2.7
6 abcd Corg 5.8
As see below, there are many functions for joining data tables.
Full joins
The full_join() function allows to make a full join between two arrays. It will keep all the data of each table, at the risk of creating gaps (NA) when some information is not present in one or the other table.
full_join(Tbl_Site, Tbl_Result, by = "Site")
# A tibble: 7 x 5
Site Landuse Area Measure Value
<chr> <chr> <dbl> <chr> <dbl>
1 356a Forest 12 Corg 3.5
2 da4b Forest 30 pH 5.8
3 da4b Forest 30 Corg 2.7
4 77de Pasture 8 <NA> NA
5 1b64 Wetland 17 pH 7
6 1b64 Wetland 17 pH 7.2
7 abcd <NA> NA Corg 5.8
The by= argument is used to specify on which variable to join. In our example, Site is used as the common key to both arrays.
Left joins
The left_join() function makes a left join. Which means on x, the table on the left. It will keep all the data from x and add the data from the right table (y) without incorporating data that does not match.
left_join(Tbl_Site, Tbl_Result, by = "Site")
# A tibble: 6 x 5
Site Landuse Area Measure Value
<chr> <chr> <dbl> <chr> <dbl>
1 356a Forest 12 Corg 3.5
2 da4b Forest 30 pH 5.8
3 da4b Forest 30 Corg 2.7
4 77de Pasture 8 <NA> NA
5 1b64 Wetland 17 pH 7
6 1b64 Wetland 17 pH 7.2
Note that the new table does not contain the abcd site, not present in x (Tbl_Site).
Right joins
In the same way the right_join() function makes a right join (on y).
right_join(Tbl_Site, Tbl_Result, by = "Site")
# A tibble: 6 x 5
Site Landuse Area Measure Value
<chr> <chr> <dbl> <chr> <dbl>
1 356a Forest 12 Corg 3.5
2 da4b Forest 30 pH 5.8
3 da4b Forest 30 Corg 2.7
4 1b64 Wetland 17 pH 7
5 1b64 Wetland 17 pH 7.2
6 abcd <NA> NA Corg 5.8
Note that the new table does not contain the 77de site, not present in y (Tbl_Result).
Filtering joins
The semi_join and anti_join functions allow to filter data table according to another.
semi_join will return all lines in x for which there is a match in y. Only the columns of x will be returned.
semi_join(Tbl_Site, Tbl_Result, by = "Site")
# A tibble: 3 x 3
Site Landuse Area
<chr> <chr> <dbl>
1 356a Forest 12
2 da4b Forest 30
3 1b64 Wetland 17
In contrast, anti_join will return all lines in x for which there is no match in y. Only the columns of x will be returned.
anti_join(Tbl_Site, Tbl_Result, by = "Site")
# A tibble: 1 x 3
Site Landuse Area
<chr> <chr> <dbl>
1 77de Pasture 8
Bind data table
When the structure of the data tables is similar, it is sometimes preferable to bind these tables to form a single one.
In this example, we will use land-use data collected in 1970 and 2020 on the same site. In 2020, the areas of each site have been added.
Tbl_Site1970 <- tibble(
Site = c("356a", "da4b", "77de", "1b64"),
Landuse = c("Crop", "Pasture", "Pasture", "Wetland")
)
Tbl_Site2020 <- tibble(
Site = c("356a", "da4b", "77de", "1b64"),
Landuse = c("Forest", "Forest", "Pasture", "Crop"),
Area = c(12, 30, 8, 17)
)
Tbl_Site1970
# A tibble: 4 x 2
Site Landuse
<chr> <chr>
1 356a Crop
2 da4b Pasture
3 77de Pasture
4 1b64 Wetland
Tbl_Site2020
# A tibble: 4 x 3
Site Landuse Area
<chr> <chr> <dbl>
1 356a Forest 12
2 da4b Forest 30
3 77de Pasture 8
4 1b64 Crop 17
There are two ways to do this:
By columns
With bind_cols, the y table is inserted to the right of the x table. The number of rows in x and y must be the identical. If column names are repeated the function adds a number behind the column name of y.
In most cases, a join is preferable…
bind_cols(Tbl_Site1970, Tbl_Site2020)
New names:
* Site -> Site...1
* Landuse -> Landuse...2
* Site -> Site...3
* Landuse -> Landuse...4
# A tibble: 4 x 5
Site...1 Landuse...2 Site...3 Landuse...4 Area
<chr> <chr> <chr> <chr> <dbl>
1 356a Crop 356a Forest 12
2 da4b Pasture da4b Forest 30
3 77de Pasture 77de Pasture 8
4 1b64 Wetland 1b64 Crop 17
Sometimes it is useful to extract a variable from a data table without importing the initial table:
bind_cols(Tbl_Site1970, select(Tbl_Site2020, Area))
# A tibble: 4 x 3
Site Landuse Area
<chr> <chr> <dbl>
1 356a Crop 12
2 da4b Pasture 30
3 77de Pasture 8
4 1b64 Wetland 17
Warning: Both tables must be ordered in the same way.
By rows
With bind_rows, the y table is inserted under the x table.
bind_rows(Tbl_Site1970, Tbl_Site2020)
# A tibble: 8 x 3
Site Landuse Area
<chr> <chr> <dbl>
1 356a Crop NA
2 da4b Pasture NA
3 77de Pasture NA
4 1b64 Wetland NA
5 356a Forest 12
6 da4b Forest 30
7 77de Pasture 8
8 1b64 Crop 17
Note that not all columns need to be present in both tables. In this case, NAs are generated.
The argument .id is used to identify which table the data comes from by creating a new column containing the id. It is possible to choose the name of the column containing the id and the identifier of each table.
bind_rows("1970" = Tbl_Site1970, "2020" = Tbl_Site2020, .id = "Year")
# A tibble: 8 x 4
Year Site Landuse Area
<chr> <chr> <chr> <dbl>
1 1970 356a Crop NA
2 1970 da4b Pasture NA
3 1970 77de Pasture NA
4 1970 1b64 Wetland NA
5 2020 356a Forest 12
6 2020 da4b Forest 30
7 2020 77de Pasture 8
8 2020 1b64 Crop 17
Manipulation of NA
In a data table, missing values (NA) are often a source of problems and difficulties.
To avoid problems, some machines use a specific code to signify missing or outlier data, for example 9999 or -9999. Sometimes it is when entering our data that we use a code, for example "NoData".
In a case like this, to perform analyses we may want to:
Convert values to NA.
The function na_if(x, y) is used to convert the values y contained in x to NA.
Here is an example:
Tbl_Temperature <- tibble(Site = c("A", "A", "A", "B", "B", "B"),
Month = c("January", "NoData", "September", "January", "June", "September"),
Temp = c(-12, 16, 13, 9999, 19, 15 ))
Tbl_Temperature
# A tibble: 6 x 3
Site Month Temp
<chr> <chr> <dbl>
1 A January -12
2 A NoData 16
3 A September 13
4 B January 9999
5 B June 19
6 B September 15
One value is missing in the Month variable (NoData) and one value is identified as an outlier in the Temp variable. We then replace these two values by NA as follow:
Tbl_TempNA <- Tbl_Temperature %>%
mutate(Month = na_if(Month, "NoData")) %>%
mutate(Temp = na_if(Temp, 9999))
Tbl_TempNA
# A tibble: 6 x 3
Site Month Temp
<chr> <chr> <dbl>
1 A January -12
2 A <NA> 16
3 A September 13
4 B January NA
5 B June 19
6 B September 15
Replace NA
The replace_na function allows you to replace the NA with one or more other values.
Tbl_TempNA
# A tibble: 6 x 3
Site Month Temp
<chr> <chr> <dbl>
1 A January -12
2 A <NA> 16
3 A September 13
4 B January NA
5 B June 19
6 B September 15
Tbl_TempNA %>%
mutate(Month = replace_na(Month, "June")) %>%
mutate(Temp = replace_na(Temp, -17))
# A tibble: 6 x 3
Site Month Temp
<chr> <chr> <dbl>
1 A January -12
2 A June 16
3 A September 13
4 B January -17
5 B June 19
6 B September 15
or :
Tbl_TempNA %>%
replace_na(list(Month = "June", Temp = -17))
# A tibble: 6 x 3
Site Month Temp
<chr> <chr> <dbl>
1 A January -12
2 A June 16
3 A September 13
4 B January -17
5 B June 19
6 B September 15
Practice
Table formatting
Transform the following data sets into a table ready for analysis, considering that the unit of observation is the season.
Tbl_Meteo consists of the average annual weather conditions.
Tbl_Weather <- tribble(
~State, ~Temp, ~Rain,
"Quebec", 15, 300,
"Ontario", 17, 280,
"Manitoba", 12, 360
)
Tbl_Obs consists of wildlife observations (positive/total).
Tbl_Obs <- tribble(
~State, ~Spring, ~Fall,
"Quebec", "22/30", "10/12",
"Ontario", "18/50", "3/4"
)
The final table should look like this:
# A tibble: 4 x 6
State Temp Rain Season Obs_posi Obs_nega
<chr> <dbl> <dbl> <chr> <int> <int>
1 Quebec 15 300 Spring 22 8
2 Quebec 15 300 Fall 10 2
3 Ontario 17 280 Spring 18 32
4 Ontario 17 280 Fall 3 1
Useful links
This workshop is largely inspired by free documents and website.
Thanks to Charles Martin PhD student at UQTR for his help.
Here are some useful links to continue:
Your favorite site:
- https://rive-numeri-lab.github.io/
Safe memory help for the tidyr package:
- https://tidyr.tidyverse.org/
More generally and to complete on the tidyverse:
- https://www.tidyverse.org/