Classe de maître avec Guillaume Blanchet
Avril 2022
Ce code permet de reproduire l’analyse de données improvisée par Guillaume Blanchet lors de sa participation à l’activité Classe de Maître du Numérilab.
Cet atelier vous est offert à titre de divertissement, mais il ne devrait en aucun cas servir de référence pour la mise en place de solutions d’analyse de données dans le cadre d’un vrai projet.
Bonne lecture!
Ouvrir les données
sp <- readRDS("sp2015.RDS")
env <- readRDS("env2015.RDS")
library(mapview)
## Warning: le package 'mapview' a été compilé avec la version R 4.1.3
mapview(sp)
# mapview(env) # Même que sp (on vient de le regarder !)
head(env)
## Simple feature collection with 6 features and 8 fields
## Geometry type: POINT
## Dimension: XY
## Bounding box: xmin: 1115906 ymin: 447007 xmax: 1255002 ymax: 500183
## Projected CRS: NAD83 / MTQ Lambert
## surface.temperature bottom.temperature bottom.salinity seal ice
## 5947 17.8 5.1 29.9 67.774815 126
## 5948 15.7 0.9 32.1 13.516211 126
## 5949 15.8 0.9 31.8 26.233578 126
## 5950 15.2 1.0 32.3 2.117578 122
## 5951 15.2 0.9 32.2 5.334104 122
## 5952 13.9 1.3 32.7 3.299375 122
## slope coarsness surfaceBottomTempDiff geometry
## 5947 0.1209286 0.6952367 12.7 POINT (1115906 447007)
## 5948 0.1679912 2.4392456 14.8 POINT (1186868 469659.8)
## 5949 0.2088341 3.0286032 14.9 POINT (1201008 469630.5)
## 5950 0.1394752 0.1658933 14.2 POINT (1209888 492226.4)
## 5951 0.3368716 6.1529986 14.3 POINT (1231779 491539.2)
## 5952 0.2605593 5.3716641 12.6 POINT (1255002 500183)
Question 1 - Relation esp - env
library(sf)
## Linking to GEOS 3.9.1, GDAL 3.2.1, PROJ 7.2.1
library(vegan)
## Warning: le package 'vegan' a été compilé avec la version R 4.1.2
## Le chargement a nécessité le package : permute
## Warning: le package 'permute' a été compilé avec la version R 4.1.2
## Le chargement a nécessité le package : lattice
## This is vegan 2.5-7
# Retirer géométrie
spDat <- st_drop_geometry(sp)
envDat <- st_drop_geometry(env)
# Ordination canonique
spHell <- decostand(spDat, method = "hellinger")
RDA <- rda(spHell, scale(envDat))
# summary(RDA)
# Graph brute !
plot(RDA, scaling = 2) # Relation entre les flèches
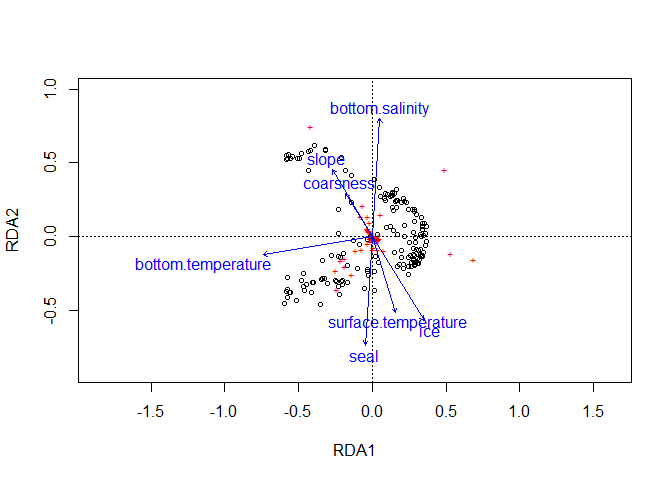
plot(RDA, scaling = 1) # Relation entre les sites
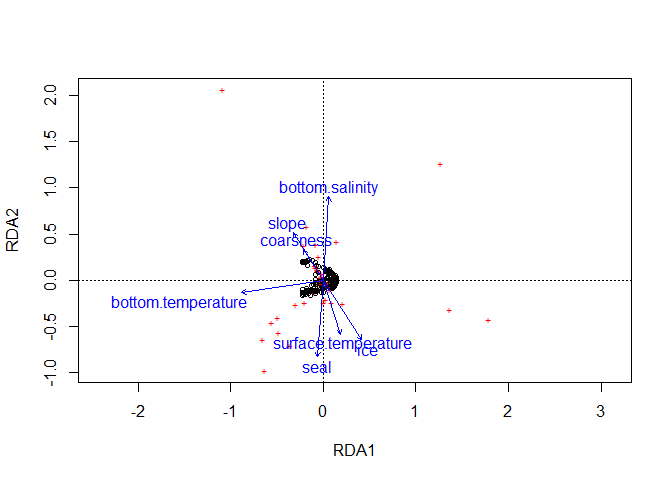
#---------------------
# Graph un peu mieux !
#---------------------
# Importance Axes
poidsAxe <- eigenvals(RDA)/ sum(eigenvals(RDA))
# position espèces dans le biplot
spAxe <- scores(RDA,
choices = 1:2,
display = "species",
scaling = 2)
# position variables environmentale dans le biplot
envAxe <- scores(RDA,
choices = 1:2,
display = "bp",
scaling = 2)
# position sites dans le biplot
siteAxe <- scores(RDA,
choices = 1:2,
display = "sites",
scaling = 2)
# Base du graph
plot(RDA,
xlab = paste0("Axe 1 - ",round(poidsAxe[1],4) * 100,"%"),
ylab = paste0("Axe 2 - ",round(poidsAxe[2],4) * 100,"%"),
type = "n",
las = 1,
scaling = 2)
# site
points(siteAxe, pch = 19, cex = 0.5)
# sp
arrows(x0 = 0,
y0 = 0,
x1 = spAxe[,1],
y1 = spAxe[,2],
angle = 10,
length = 0.1,
col = "red")
text(spAxe,
labels = rownames(spAxe),
col = "red",
cex = 0.4)
# env
arrows(x0 = 0,
y0 = 0,
x1 = envAxe[,1],
y1 = envAxe[,2],
angle = 10,
length = 0.1,
col = "blue")
text(envAxe,
labels = rownames(envAxe),
col = "blue",
cex = 0.6)
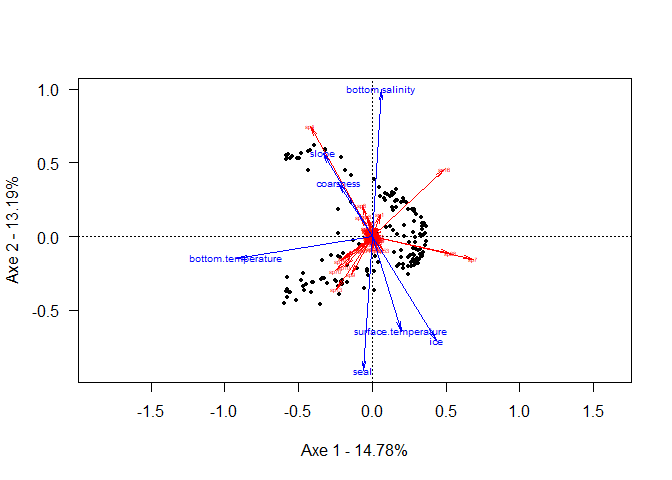
Relation espace - env pour la communauté
Moran’s eigenvector maps pour variables spatiales
library(adespatial)
## Warning: le package 'adespatial' a été compilé avec la version R 4.1.3
## Registered S3 methods overwritten by 'adegraphics':
## method from
## biplot.dudi ade4
## kplot.foucart ade4
## kplot.mcoa ade4
## kplot.mfa ade4
## kplot.pta ade4
## kplot.sepan ade4
## kplot.statis ade4
## scatter.coa ade4
## scatter.dudi ade4
## scatter.nipals ade4
## scatter.pco ade4
## score.acm ade4
## score.mix ade4
## score.pca ade4
## screeplot.dudi ade4
## Registered S3 method overwritten by 'spdep':
## method from
## plot.mst ape
## Registered S3 methods overwritten by 'adespatial':
## method from
## plot.multispati adegraphics
## print.multispati ade4
## summary.multispati ade4
#?mem #Pour les MEMs super spécifique
# pcnm - Général mais pas optimal (bon pour l'illustration)
PCNM <- pcnm(dist(st_coordinates(sp))) # dans vegan
# calcul R2a
vp <- varpart(spHell,
PCNM$vectors,
PCNM$vectors)
## Warning: collinearity detected in cbind(X1,X2): mm = 184, m = 92
# Forward selection sur les variables spatiales (PCNM)
fs <- forward.sel(Y = spHell,
X = PCNM$vectors,
adjR2thresh = vp$part$fract[1,3],
alpha = 0.06) # dans adespatial
## Testing variable 1
## Testing variable 2
## Testing variable 3
## Testing variable 4
## Testing variable 5
## Testing variable 6
## Testing variable 7
## Testing variable 8
## Testing variable 9
## Testing variable 10
## Testing variable 11
## Testing variable 12
## Testing variable 13
## Testing variable 14
## Testing variable 15
## Testing variable 16
## Testing variable 17
## Testing variable 18
## Testing variable 19
## Testing variable 20
## Testing variable 21
## Testing variable 22
## Testing variable 23
## Testing variable 24
## Testing variable 25
## Testing variable 26
## Testing variable 27
## Testing variable 28
## Testing variable 29
## Testing variable 30
## Testing variable 31
## Testing variable 32
## Procedure stopped (alpha criteria): pvalue for variable 32 is 0.075000 (> 0.060000)
fs
## variables order R2 R2Cum AdjR2Cum F pvalue
## 1 PCNM5 5 0.045437491 0.04543749 0.03943395 7.568453 0.001
## 2 PCNM2 2 0.039319304 0.08475679 0.07317144 6.787759 0.001
## 3 PCNM7 7 0.028906271 0.11366307 0.09672669 5.120270 0.001
## 4 PCNM13 13 0.026701675 0.14036474 0.11832281 4.845615 0.001
## 5 PCNM8 8 0.024751000 0.16511574 0.13818399 4.595134 0.001
## 6 PCNM3 3 0.021682337 0.18679808 0.15511489 4.106089 0.001
## 7 PCNM19 19 0.016983719 0.20378180 0.16735351 3.263564 0.005
## 8 PCNM6 6 0.016047654 0.21982945 0.17876784 3.126552 0.002
## 9 PCNM36 36 0.015528774 0.23535823 0.18978355 3.066593 0.001
## 10 PCNM20 20 0.014508823 0.24986705 0.19985818 2.901250 0.005
## 11 PCNM11 11 0.013786430 0.26365348 0.20929233 2.789689 0.004
## 12 PCNM1 1 0.013695869 0.27734935 0.21875605 2.804936 0.007
## 13 PCNM4 4 0.013591923 0.29094127 0.22823540 2.817838 0.009
## 14 PCNM92 92 0.013477706 0.30441898 0.23771943 2.828923 0.004
## 15 PCNM10 10 0.013461742 0.31788072 0.24731666 2.861600 0.002
## 16 PCNM21 21 0.013102116 0.33098283 0.25664759 2.820114 0.009
## 17 PCNM24 24 0.012770203 0.34375304 0.26573766 2.782701 0.004
## 18 PCNM15 15 0.012705285 0.35645832 0.27488262 2.803471 0.005
## 19 PCNM17 17 0.011332287 0.36779061 0.28259927 2.527410 0.006
## 20 PCNM33 33 0.010810143 0.37860075 0.28982943 2.435503 0.014
## 21 PCNM9 9 0.010452063 0.38905282 0.29675144 2.378007 0.022
## 22 PCNM23 23 0.010250380 0.39930320 0.30353994 2.354853 0.015
## 23 PCNM12 12 0.010102585 0.40940578 0.31025493 2.343494 0.014
## 24 PCNM16 16 0.009679705 0.41908549 0.31657116 2.266151 0.017
## 25 PCNM18 18 0.009384354 0.42846984 0.32263092 2.216660 0.028
## 26 PCNM14 14 0.008966696 0.43743654 0.32828243 2.135825 0.023
## 27 PCNM30 30 0.008964658 0.44640119 0.33401647 2.153725 0.028
## 28 PCNM75 75 0.008769852 0.45517105 0.33960127 2.124741 0.034
## 29 PCNM26 26 0.008554484 0.46372553 0.34500828 2.089671 0.029
## 30 PCNM28 28 0.007724472 0.47145000 0.34947693 1.899880 0.047
## 31 PCNM72 72 0.007523453 0.47897346 0.35376553 1.862718 0.050
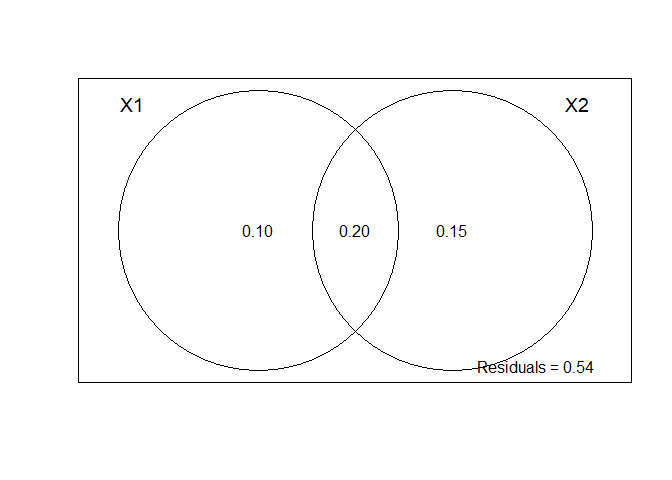
# Partition de la variation
vpRes <- varpart(spHell,
scale(envDat),
PCNM$vectors[,fs$order])
## Warning: collinearity detected in X1: mm = 8, m = 7
## Warning: collinearity detected in cbind(X1,X2): mm = 39, m = 38
vpRes
##
## Partition of variance in RDA
##
## Call: varpart(Y = spHell, X = scale(envDat), PCNM$vectors[, fs$order])
##
## Explanatory tables:
## X1: scale(envDat)
## X2: PCNM$vectors[, fs$order]
##
## No. of explanatory tables: 2
## Total variation (SS): 100.64
## Variance: 0.629
## No. of observations: 161
##
## Partition table:
## Df R.squared Adj.R.squared Testable
## [a+b] = X1 7 0.33639 0.30603 TRUE
## [b+c] = X2 31 0.47897 0.35377 TRUE
## [a+b+c] = X1+X2 38 0.58501 0.45575 TRUE
## Individual fractions
## [a] = X1|X2 7 0.10199 TRUE
## [b] 0 0.20404 FALSE
## [c] = X2|X1 31 0.14972 TRUE
## [d] = Residuals 0.54425 FALSE
## ---
## Use function 'rda' to test significance of fractions of interest
plot(vpRes)